
Donald Rio
Biochemistry of P element transposase, the mechanism of transposition and activity of P element-related THAP9 genes in humans and zebrafish:
The 87kD P element-encoded transposase protein is required to catalyze P element transposition and belongs to a large polynucleotidyl transferase superfamily, that includes RNaseH, RuvC, retroviral integrases, transposases and the argonaute proteins. Biochemical studies using the purified protein revealed that guanosine triphosphate (GTP) is an essential cofactor for the reaction. Current studies involve the use of imaging methods, including atomic force microscopy (AFM), electron microscopy (EM) and total internal reflection fluorescence microscopy (TIRFM) to understand the role(s) that GTP plays in transposition, the detailed reaction pathway and how this cofactor modulates the assembly and activity of transposase on DNA. We are also interested in using X-ray crystallography to study P element transposase bound to DNA, and initially in collaboration with James Berger’s lab, have solved the structure of the N-terminal THAP DNA binding domain (DmTHAP bound to DNA; see figure below). The C2CH zinc-coordinating THAP DNA binding domain is found in many animal genomes in >300 genes. Interestingly, the THAP9 gene in humans and zebrafish shares extensive sequence homology to the Drosophila P element transposase and we have shown that the THAP9 gene encodes an active DNA transposase. We are using genome-wide methods, ChIP-seq, to find locations that THAP9 binds in the human genome.
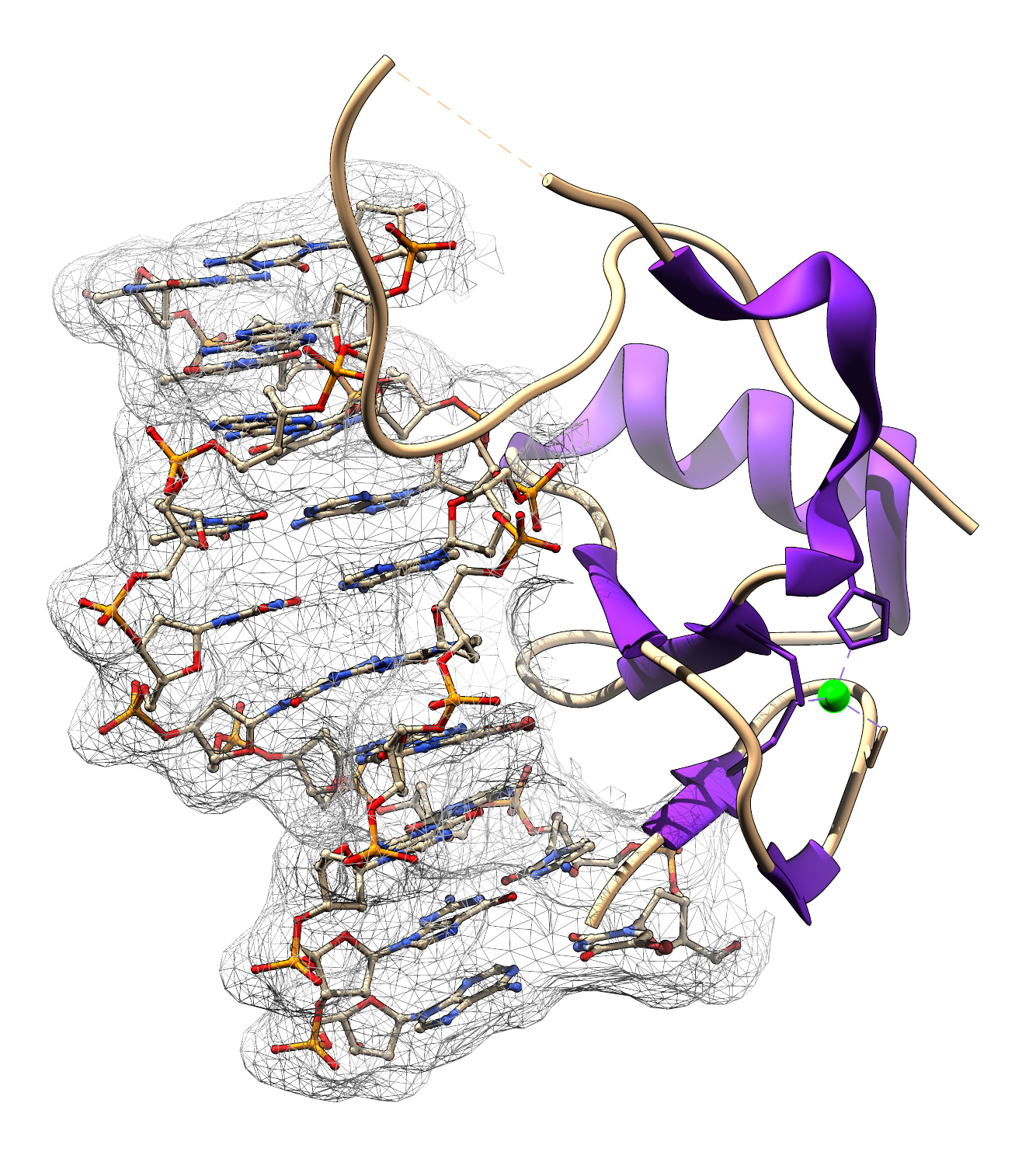 Structure of DmTHAP-DNA complex.
Structure of DmTHAP-DNA complex.
a) Overall structure of the DmTHAP-DNA complex. Experimental electron density map of the DNA (blue mesh) is contoured at 1.5σ. DmTHAP is shown as a ribbon diagram with the βαβ motif highlighted in magenta. Zinc is shown as a green sphere for clarity.
b) Base-specific interactions in the major and minor groove. Interacting amino acids are shown as magenta sticks; DNA is shown in surface representation; zinc-coordinating residues are in green.
Biochemistry, genome-wide and systems-level analysis of alternative pre-mRNA splicing and splicing silencer function in Drosophila and humans.
The P element transposon pre-mRNA undergoes tissue-specific splicing and we showed that regulation of the third P element intron (IVS3) involves RNA binding proteins (PSI, hrp48, hrp36 and hrp38) that recognize an exonic splicing silencer (ESS) regulatory RNA element in the 5′ exon adjacent to IVS3, resulting in splicing inhibition. This RNA silencer element binds the splicing repressor protein PSI (P element somatic inhibitor) protein which directly interacts with U1 snRNP and modulates U1 snRNP binding to specific sites on the pre-mRNA. We are using bioinformatic and genomic (microarray and mRNA-seq) approaches to identify and characterize new splicing silencers that use the hrp48 and PSI factors in cellular genes. We are employing biochemical reconstitution and purification, whole-genome tiling microarrays and mass spectrometry to identify cellular mRNAs and proteins present in RNP particles containing PSI, hrp48 and other splicing factors. We are also using RNA tagging methods to purify and analyze the protein composition of RNP and splicing silencer complexes assembled in vitro and in vivo. We are interested in whether the incorporation of hnRNP proteins into RNP particles on nascent pre-mRNAs might generate a “code” that specifies how pre-mRNAs are processed. Another project involves a Drosophila paralog of the splicing factor U2 snRNP auxiliary factor (U2AF) that appears to act as a splicing silencer. We are investigating the roles of the microRNA pathway gene, Argonaute-2 (Ago-2), in the nucleus. Using mutants, RNA interference, CLIP-seq and RNA-seq our data indicate that Ago-2 determines genome-wide patterns of alternative pre-mRNA splicing and Polycomb-group (PcG)-mediated transcriptional repression in Drosophila. Finally, as part of the newly formed Center for RNA Systems Biology (Jamie Cate, PI), we embarking on an investigation of transcriptome structure in humans using chemical probing and high-throughput sequencing (SHAPE-seq/DMS-seq) and the role of the RNA chaperone proteins, hnRNA A1 and DDX5 in alternative pre-mRNA splicing decisions and RNA structural alterations, with the goal of linking specific RNA binding or structural features to normal or aberrant splicing patterns.

